Research
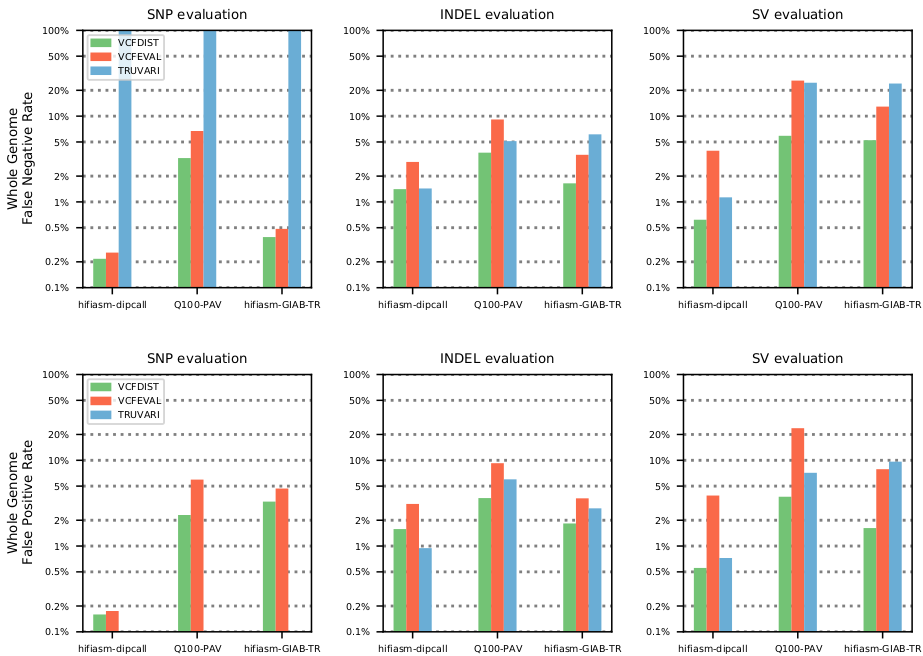
Jointly benchmarking small and structural variant calls with vcfdist
Advisors: Satish Narayanasamy, Justin Zook, James HoltMar 2023 - Jan 2024
In this work, I extended my existing small variant calling evaluator vcfdist to handle evaluation of structural variant calls as well. This involved significant restructuring and optimization to handle increased memory and computational demands. vcfdist is now the only tool to perform sophisticated whole-genome analysis of phased small and structural variant calls. The figure above shows that it also reduces measured false negative and false positive rates in comparison to existing small and structural variant calling evaluators, vcfeval and Truvari.Paper: Jointly benchmarking small and structural variant calls with vcfdist
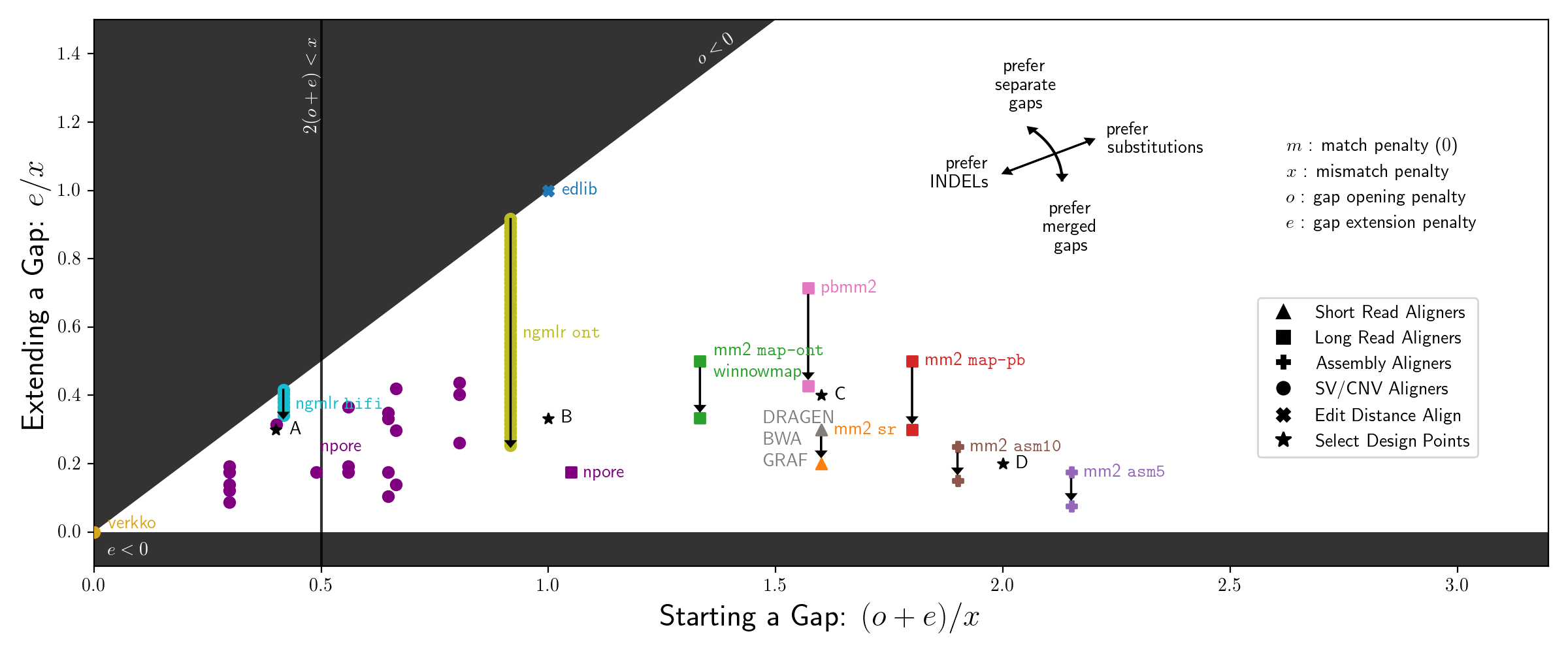
vcfdist: Accurately benchmarking small variant calls in human genomes
Advisor: Satish NarayanasamyFeb 2022 - Feb 2023
vcfdist is a distance-based small variant calling evaluator that:
- standardizes query and truth VCF variants to a consistent representation
- discovers long-range variant representation dependencies using a novel clustering algorithm
- requires local phasing of both input VCFs and enforces correct local phasing of variants
- gives partial credit to variant calls which are mostly correct
Paper: vcfdist: Accurately benchmarking small variant calls in human genomes
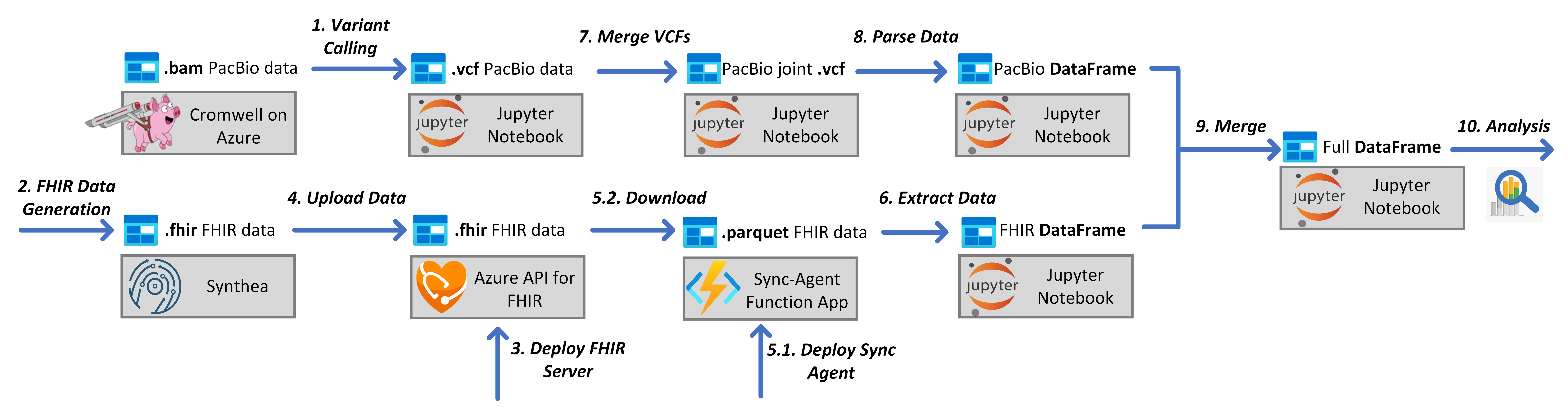
A cloud-based pipeline for analysis of FHIR and long-read data
Advisor: Erdal CosgunJune - Aug 2022
During my MSR internship I developed a secure and scalable cloud-based pipeline for working with both PacBio sequencing data and clinical FHIR data, from initial data through tertiary analysis. The electronic health records are stored in FHIR -- Fast Healthcare Interoperability Resource -- format, the current leading standard for health care data exchange. For the genomic data, we perform variant calling on long read PacBio HiFi data using Cromwell on Azure. Both data formats are parsed, processed, and merged in a single scalable pipeline which securely performs tertiary analyses using cloud-based Jupyter notebooks. We then demonstrate three example downstream applications: exporting patient information to a database, clustering patients, and performing a simple pharmacogenomic study.
Paper: A cloud-based pipeline for analysis of FHIR and long-read data
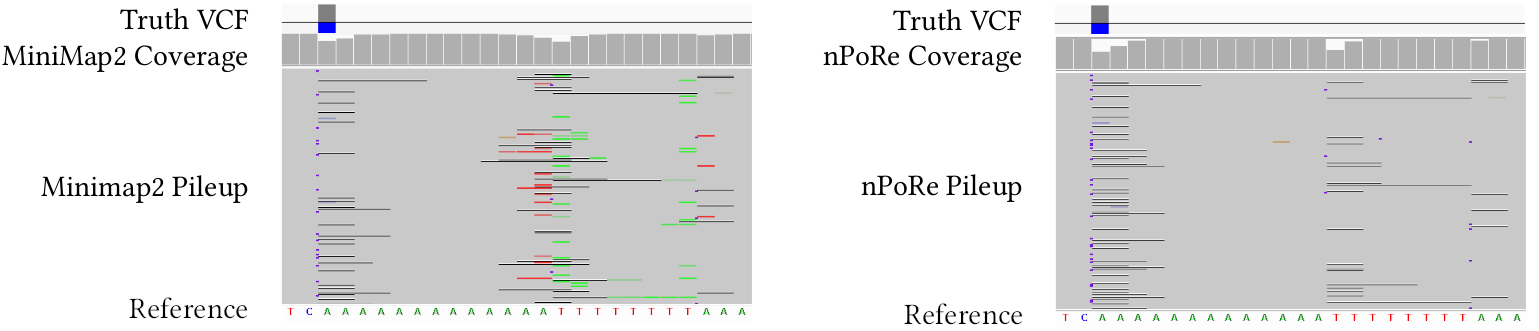
nPoRe: n-Polymer Realigner for improved pileup variant calling
Advisor: Satish NarayanasamyMay 2021 - Feb 2022
nPoRe is a nanopore sequencing read realigner which recalculates each read's fine-grained alignment in order to more accurately align ''n-polymers'' such as homopolymers (n=1) and tandem repeats (2 <= n <= 6). In other words, given an input BAM, it adjusts each read's CIGAR string to more accurately model the most likely sequencing errors and actual variants. Traditional affine gap penalties are context-agnostic, and do not model the higher likelihood of INDELs in low-complexity regions (particularly n-polymers), leading to poor or inconsistent alignments. We find that npore improves pileup concordance across reads and results in slightly better variant calling performance.
Pre-Print: nPoRe: n-Polymer Realigner for improved pileup-based variant calling
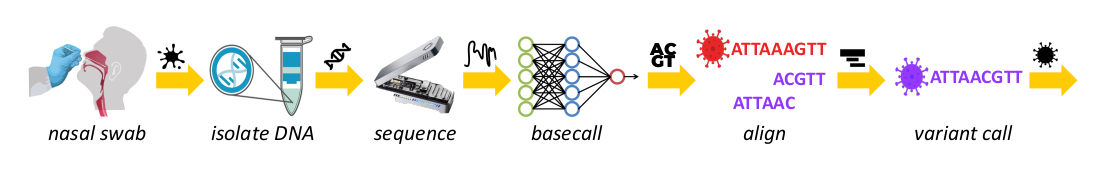
SquiggleFilter: An Accelerator for Portable Virus Detection
Advisor: Satish NarayanasamySep 2019 - Dec 2020
Nanopore sequencing is a relatively new technology which enables portable genome sequencing, including strain-level virus detection. The vast majority of DNA in a typical spit sample is non-viral. Nanopore sequencers provide a unique feature called Read Until, which can eject non-viral DNA strands in real time to increase the proportion of viral DNA sequenced. This approach comes with several computational challenges, and in this work we propose replacing the traditional pipeline of basecalling and alignment with a hardware accelerated squiggle-level read filter. Our resulting SquiggleFilter is an efficient high thoughput and low latency non-viral read filter capable of keeping pace with the next generation of sequencing devices.
Paper: SquiggleFilter: An Accelerator for Portable Virus Detection
Conference: MICRO 2021
Badges:

 Awards: IEEE MICRO 2022 Top Picks Honorable Mention
Awards: IEEE MICRO 2022 Top Picks Honorable Mention

Hypervisor vTPM Integration
Advisors: Paul Mogren and Rich TurnerMay - Aug 2019
During my internship at Assured Information Security (AIS) I worked with Danika Gaviola and Jason Rising on integrating a virtual Trusted Platform Module (vTPM) into OpenXT in order to provide virtual machines (VMs) the same security guarantees as a physical TPM would provide. A TPM is a secure cryptoprocessor most frequently used to verify machine state at each stage of the boot process. OpenXT is a security-enhanced fork of Xen which uses QEMU internally for peripheral device emulation. In addition to successfully providing TPM 2.0 for Linux VMs in OpenXT, we modified the Xen Project so that it could use TPM 2.0 and provide TPMs to Windows virtual machines.
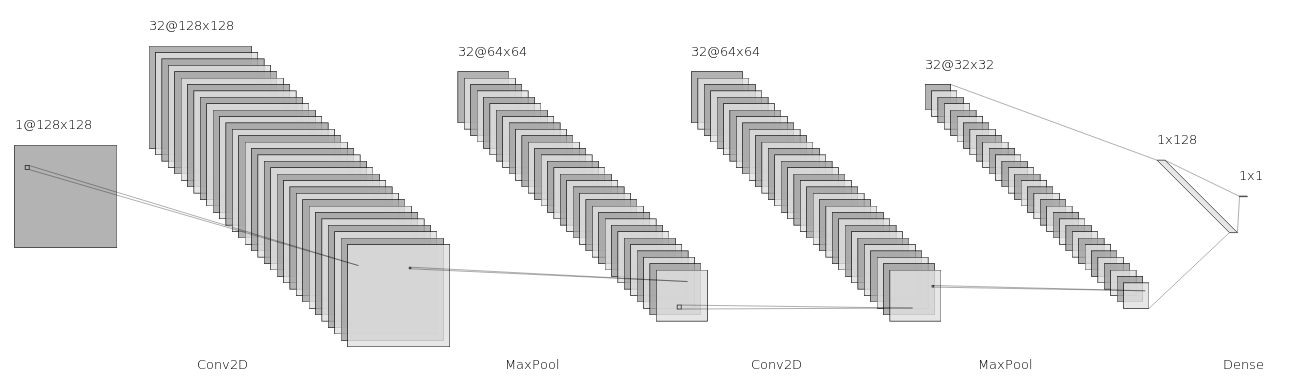
Using ConvNets for Robust Swipe Pressure Detection
Advisors: Sean and Natasha BanerjeeSep - Dec 2018
This research constitutes my Clarkson Honors Thesis, which extends my previous work on using thermal cameras to reliably detect swipe pressure. I explored using convolutional neural networks to perform action classification, and found that the best results were obtained by training ConvNets on video frames with the user's hand removed, using filtering techniques designed previously.
Honors Thesis: Detection of Swipe Pressure using a Thermal Camera and ConvNets for Natural Surface Interaction
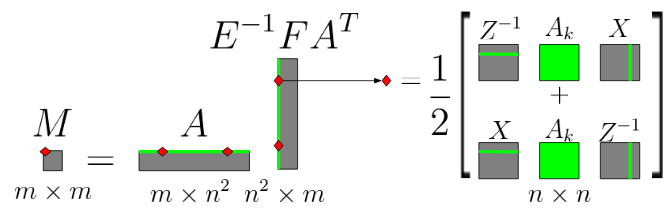
Parallel Semidefinite Optimization
Advisor: Daniel ThuerckMay - Aug 2018
During my internship at TU Darmstadt I worked on extending an existing solver for linear optimization programs to perform semidefinite optimization. The most challenging obstacle I overcame involved computing the product of multiple large sparse matrices without storing any intermediate results. Each element in the resulting matrix could be computed individually due to certain matrix properties, and the process is shown in the image above.

User-Independent Detection of Swipe Pressure Using Thermal Cameras
Advisors: Sean and Natasha BanerjeeJan - May 2018
The goal of this research was to improve the capabilities of current augmented reality systems and projective touchscreens by detecting the pressure at which a user interacts with objects using a thermal camera. The main challenge in developing such a classifier was in removing the user's hand from the thermal video, which required utilizing a series of morphological filters as demonstrated above.
Paper: User-Independent Detection of Swipe Pressure using a Thermal Camera for Natural Surface Interaction Conference: MMSP 2018 Awards: Top 5% Paper

Ptychographic Image Reconstruction
Advisor: Christine SweeneyMay - Aug 2017
During my internship at Los Alamos National Laboratory I worked with an interdisciplinary team of scientists to enable their ptychographic image reconstruction algorithms to run on CPUs in addition to Los Alamos' GPU cluster. Afterwards, I greatly improved the software's ease of use and automated the installation and building processes. Lastly, I worked on implementing a parallel beam position refinement algorithm to improve image reconstruction accuracy.
Image from http://compphotolab.northwestern.edu/project/adp-automatic-differentiation-ptychography/
Optimization of MATLAB PointCloud Generation
Advisors: Sean and Natasha BanerjeeJan - May 2017
While working with the TARS research group at Clarkson, I improved the algorithms for generating a single PointCloud from our multi-Kinect system. By the end of the semester, I had decreased the total runtime required to generate a single PointCloud from 7.33 seconds to 0.178 seconds.
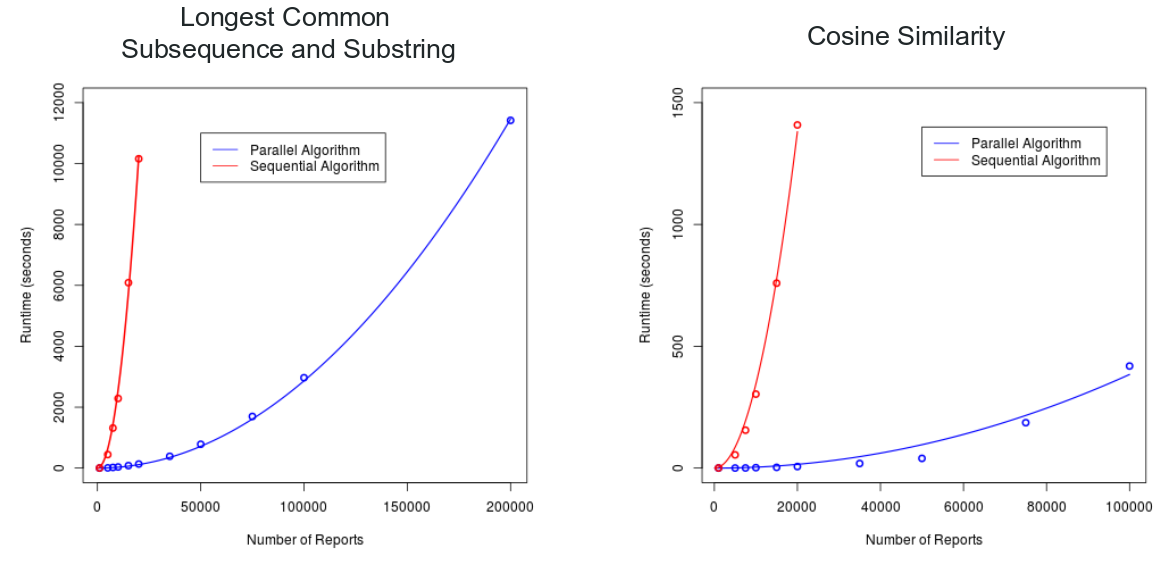
Using GPUs to Mine Large-Scale Software Problem Repositories
Adviser: Sean BanerjeeMay - July 2016
This project involved accelerating the detection of duplicate problem reports in large software problem report repositories. I implemented several basic parallel algorithms in CUDA C++ which could more quickly compare reports from massive repositories such as Bugzilla.
Paper: GPU Acceleration of Document Similarity Measures for Automated Bug Triaging Workshop: IWSF 2016 Awards: Best Poster Presentation in EE & CS, Clarkson University Symposium on Undergraduate Research Experiences